By Andrew Little, Thematic Research Analyst at Global X ETFs
In early January, researchers identified an irregular, lethal pneumonia spreading through China as the disease we now know as COVID-19. Global efforts to identify, treat, and limit the spread of the disease mobilized unprecedented resources toward Genomics, a cutting-edge field of biology with especially valuable medical applications.
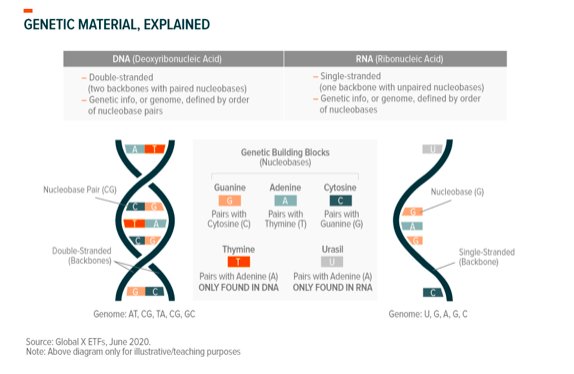
Genomics is the study of an organism’s complete set of genetic information, or genome. Using ourselves for context, our DNA contains our genetic information and the complete set of our DNA is our genome. Every living thing has its own genome. Viruses, which are not alive, also have their own genomes. Genomic sub-sciences like genomic sequencing and computational genomics give us insight into genomes’ functionality, while genetic therapies and gene editing seek to use that information for medical or other purposes.
Each of these genomic sub-sciences is helping to understand and address COVID-19 and the virus behind it, SARS-CoV-2 or ‘coronavirus.’ From the virus’ first known days to now, we can observe impactful applications of each and glimpse what this might mean for genomics moving forward.
In the following, we explore genomics’ critical role in addressing the COVID-19 pandemic at-hand and investigate how the science could transform therapeutics and the healthcare industry as a whole.
Genomic Sequencing and Computation: Discovering, Diagnosing, and Tracing COVID-19
Genome sequencing reveals the genetic makeup of organisms like humans, or even bacteria, by identifying the building blocks (nucleotides) that make up genetic material (DNA) and the order in which they appear. Sequencing can also be conducted on viral genomes like that of SARS-CoV-2. Viruses are non-living strands of RNA, so these sequences look at RNA rather than DNA.
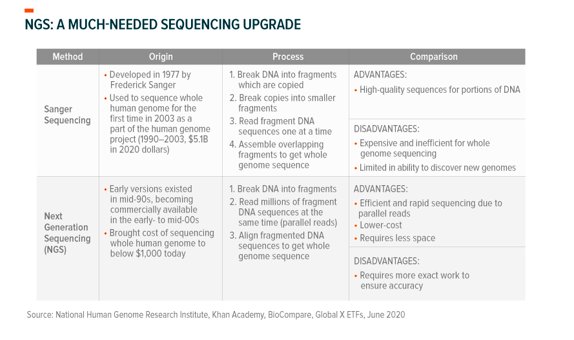
When pneumonia patients in Wuhan, China, didn’t respond to treatment, Chinese researchers sequenced genetic material from lung fluid samples and identified SARS-CoV-2 for the first time.1 In the past, scientists sequenced DNA one fragment at a time in what is known as Sanger sequencing.2 While the Sanger method was instrumental in sequencing the first human genome in 2003, its reliance on known sequences and slow throughput (speed) limit its ability to quickly identify novel pathogens.3,4 Today, there are new techniques and tools available that enable faster and more reliable whole genome sequencing.
Next generation sequencing (NGS) reads millions of DNA fragments simultaneously and requires no prior knowledge of the genome, lending itself well to identifying new pathogens.5 Researchers employed Illumina and Nanopore NGS to read SARS-CoV-2’s genome and computational genomic applications like QIAGEN’s CLC software to map and sequence it. From this, they were able to determine that the virus was a novel pathogen.6,7Just 12 days after the first public announcement of a pneumonia outbreak, they published its genome.8 This initial effort represents a new age of identifying infectious disease – for comparison, it took around 6 months to sequence the novel SARS virus in 2003.9,10 Most importantly, the initial sequencing work set the table for the progress we’ve made in the months since.
Diagnostics
Initial and subsequent sequencing work enabled the prompt development of COVID-19 tests (assays). Researchers used the first genome sequences to develop reverse transcription polymerase chain reaction (RT-PCR) tests, which are a type of molecular assay.11 RT-PCR tests look for genetic traces of a specified pathogen – in this case, the SARS-CoV-2 genome – from patient samples (nasal swab). A COVID-19 RT-PCR test works as follows:12
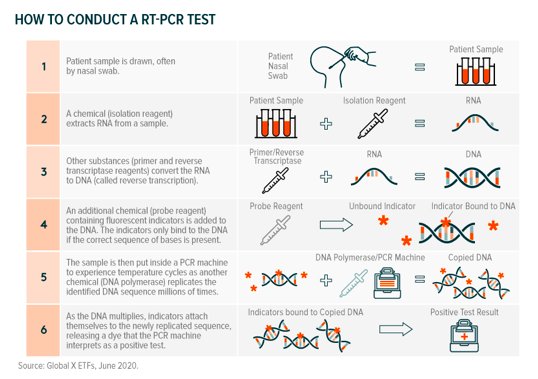
The value chain for RT-PCR assays includes PCR machine manufacturers, reagent producers, and designers of all-in-one testing kits. Hong Kong biotech company Genscript, for example, was among the first to come to market with a SARS-CoV-2 RT-PCR assay and produces the reagents used to turn RNA to DNA and indicate the presence of the virus.13,14
Agilent, a Silicon Valley-based company, similarly produces reagents, but also manufactures the Aria Mx/Dx qPCR machine which can be used in the RT-PCR process.15 Other molecular testing assays include CRISPR gene-editing based technologies, isothermal amplification, and hybridization, though 90% of tests utilized RT-PCR up to this point.16
Serology-based tests, or immunological assays, look for the presence of proteins we produce to fight infections (antibodies) in bodily fluids, indicating an immune response to the virus (pathogen).17 Different antibodies are specific to different viruses. IgM antibodies appear in subjects recently infected by the virus, while IgG antibodies appear on the tail-end and following infection.18
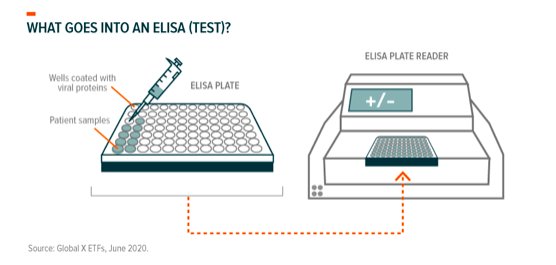
As many infected with COVID-19 are asymptomatic, IgG antibody testing can reveal whether one unknowingly fought off infection. This could prove valuable for uncovering the extent of an outbreak and possible immunity. Enzyme-linked immunoabsorbent assays (ELISA) are the primary tests available for SARS-CoV-2 antibody-based diagnoses. Typically, patient blood samples are placed in small, virus-covered wells on a plastic plate which is then fed into an ELISA plate reader.
The reader detects whether antibodies in the patient sample interact (bind) with viral proteins called antigens. Companies like Agilent, through their Biotek subsidiary, produce the instruments needed in this process, including plate readers and washers. Over a dozen other companies also produce ELISA kits and/or the components within.19,20
Additional serology-based tests for SARS-CoV-2 include lateral flow immunoassays, which are usually at-home rapid tests, and virus neutralization tests, which look to see if patients’ antibodies actually fight the virus.21 Many ELISA and lateral flow tests are subject to inaccuracy and don’t specify if present antibodies neutralize the virus. This is where neutralization tests play a part, but most of these tests require strict biosafety facilities and it’s still unknown how long neutralizing antibodies are effective.22 Genscript produces a version of this test with less biohazard risk that’s approved in Europe and Singapore, though the FDA has yet to approve it.23
Surveillance
Sequencing is also important for surveillance efforts which, similar to genome discovery, relies on the breakthrough efficiency of next generation sequencing (NGS) to read and sequence the virus’ entire genome. As discussed, NGS reads millions of DNA fragments concurrently and is considerably faster than past methods. Companies like Illumina, Pacific BioSciences, and QIAGEN enable this via their NGS platforms which include preparation kits (containing reagents and DNA fragments), sequencing machines, and analysis software.24
Researchers have added 41,000+ SARS-CoV-2 sequences to GISAID’s public genome database (as of 6/8/2020) since the first publication of SARS-CoV-2’s genome in January.25
Analysis done on sequences from earlier infections provide insight into the virus’ origin and fundamental characteristics. Of note, SARS-CoV-2’s genome shares 79.6% sequence identity with 2003’s SARS virus and 96% with a coronavirus found in bats.26 Also similar to 2003’s SARS virus, SARS-CoV-2 uses the ACE2 cell receptor to infect humans.27
From these findings, the scientific community deduced that 1) SARS-CoV-2 is entirely novel (new to humans), and 2) since it latches on to cells via the ACE2 receptor, likely came from the wild, spreading to humans either through bats or pangolins (zoonotic).28 Computational genomic analysis processed by artificial intelligence gives us further detail, assessing with high probability that the virus originated from bats.29
Subsequent genomes give us an idea of the virus’ transmission path. SARS-CoV-2 mutates at an average rate of two alterations (mutations) a month as a result of natural replication errors and natural selection.30 This means that if you sequenced the virus in a sick person today and followed the infection from person to person, the virus found in those a month later would have two differences in its sequence (on average). However, these two alterations could be entirely different between individuals indirectly infected from the same original host.31 As you can see below, it looks similar to a family tree:
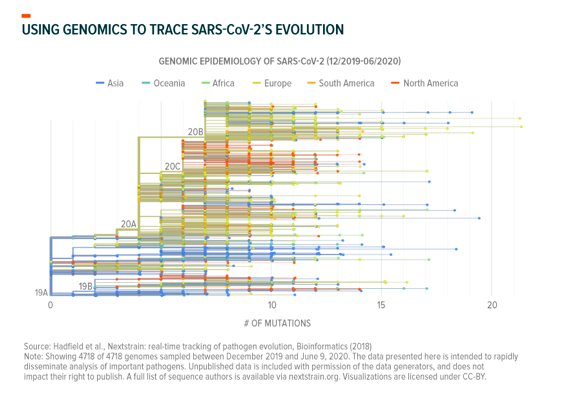
Computational biologists apply this logic at massive scale to trace transmission. They take raw data from the 41,000+ different genomes associated with different mutations, locations, and time of infection; apply AI algorithms; and map how the virus spreads. This tells them how long COVID-19 circulated in specific regions and how it got there. Viral genomes sequenced in New York City, for example, mainly follow a transmission path that appears to lead back to Europe, though many lead to other North American localities.32 Separately, researchers in Washington State recently used computational genomics to probabilistically trace their outbreak back to Hubei, China, via either repatriation amid February’s travel restrictions or through the Canadian border.33
Here’s SARS-CoV-2’s transmission path, mapped as of June 9, 2020:
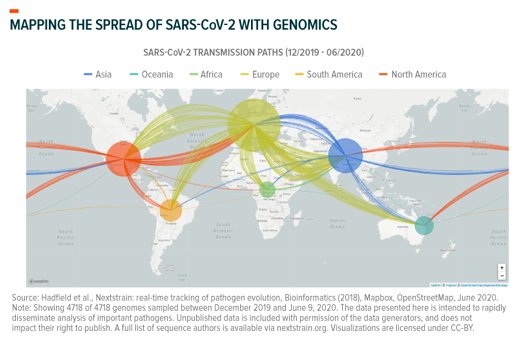
Finally, surveillance sequencing could reveal how mutations affect the virus’ behavior or treatment options. Varied studies published since January report mutations altering its contagiousness, lethality, and ability to be treated. While these types of mutations are possible and warrant study, it is important to note that many attempts at characterizing the virus to this level of detail lack peer review and are subject to criticism.34
Collaborative efforts between academics, governments, and corporations allowed diagnostics and surveillance to scale faster than previously imagined: as of June 5th, the global health community conducted over 30 million diagnostics tests in addition to the 41,000 published whole genome sequences.35 The advancements we’ve seen over the past 6 months should serve as a foundation for genomic approaches in the future, just as the many decades of work prior to the pandemic primed genomics for the spotlight it holds today.
Genomic Medicines and Their Potential to Treat and Cure COVID-19
Gene-based treatments and vaccines derived from sequencing could present our most immediate hopes of treating those afflicted and thwarting both current and future pandemics.
Treatments
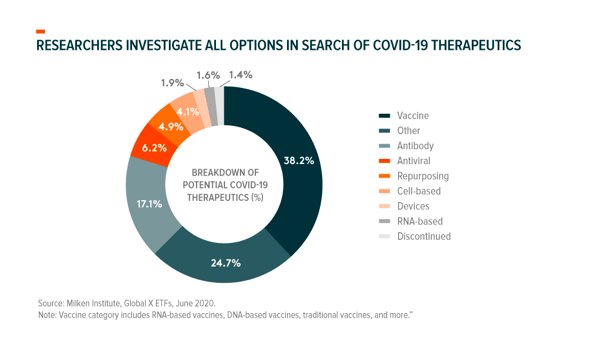
COVID-19 treatments seek to treat existing SARS-CoV-2 infections and decrease attributable mortality. As of June 16, 2020, there were 230+ different treatments under consideration globally, but only two drugs had Emergency Use Authorization (EUA) from the FDA.36 EUA doesn’t constitute a drug approval, but gives the FDA power to make an unapproved product available in times of crisis.37
Broadly, there are three ways to limit viral diseases like COVID-19: stopping the virus from entering our cells, interfering with its ability to replicate and spread within us, and minimizing the damage it does to our bodies. Yet, the first step before assessing each line of defense is understanding host to virus interactions. Computational genomic software, like QIAGEN’s Ingenuity Path Analysis (IPA) tool, process genomic data from organisms and viruses to provide an assessment of interactions between the two.38 These interactions give researchers an idea of where therapeutics can come into play.
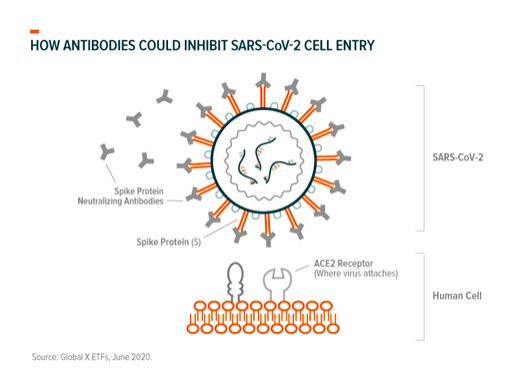
Inhibiting entry into cells: As with other coronaviruses, SARS-Cov-2 attaches to our cells the way a key enters a lock. The “keys” are the crown-shaped spike proteins (glycoprotein) found on the virus’ outer structure (see graphic) and the “locks” are the receptor proteins (ACE2) on our cells’ surface.39
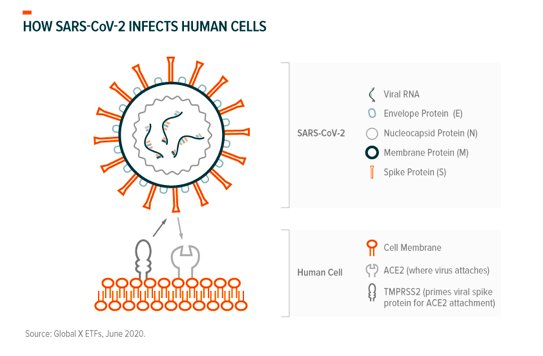
SARS-CoV-2’s genome shapes the virus’ spike proteins, in this case making them a perfect fit to bind with our ACE2 (angiotensin-converting enzyme 2) receptors where the viral genome (RNA) enters our cells through other virus-host protein interactions.40 TMPRSS2 (transmembrane protease, serine 2) is a protein our cells produce to prime spike proteins for binding and is involved in this process.41
Researchers can use this knowledge to determine which proteins or interactions can be targeted by existing drugs or compounds, that are either “off-the-shelf” or under separate investigation.42 Within the Chemical Abstract Service’s (CAS) drug/compound database, researchers identified 430 existing drugs related to ACE2 and TMPRSS2 that could be potential candidates for further vetting.43 Unfortunately, many compounds either have no effect, are effective in labs (in vitro) but not in the body (in vivo), or are dangerous for human consumption.
The other part of drug discovery is finding new compounds that “drug” proteins or interactions. Modern computing can be an effective way to do this: IBM’s Summit supercomputer recently found 77 compounds that block SARS-CoV-2’s spike protein.44There are a number of new antiviral and antibody treatments currently under consideration that could stop the virus from binding with our cells. As an example, Sorrento Therapeutics’ preclinical COVIDTRAP seeks to introduce antiviral decoy receptors that the virus could bind to before reaching our cells.45
Efforts to explore the use of antibodies in attacking SARS-CoV-2’s structural features (spike protein and shell) are also underway. Sorrento’s preclinical antibody treatments, COVI-SHIELD and COVI-GUARD, take a different approach that targets SARS-CoV-2’s spike protein via antibodies that bind with it.46 A recent study observed positive results in patients who received transfusions of blood plasma from patients who successfully fought off COVID-19 and had corresponding antibodies.47 In a similar vein, researchers are testing the efficacy of “growing” antibodies within other animals’ immune systems and transferring them to infected humans.48
Other possible approaches to inhibiting the infection of healthy cells include synthetically manipulating the ACE2 receptor, interfering with TMPRSS2-Spike protein interactions, and interfering with protein building-block (amino acids) transporters.49
Interfering with replication: After viral RNA enters our cells, its sole purpose is to replicate as many times as possible to create new viruses that can infect other cells. There are multiple processes and components that go into this:
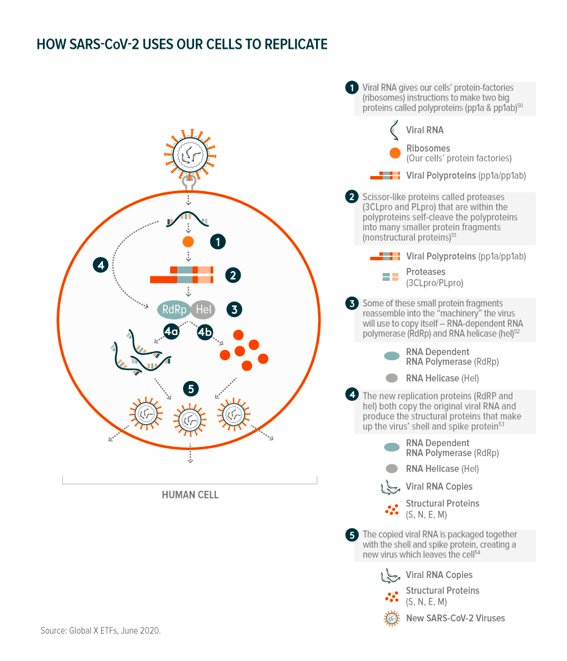
The number of viral proteins and interactions with the host cell means that there are many “druggable” pathways – CAS’ database identifies 2,937 drug candidates between the two scissor-like protease proteins (3CLpro and PLpro) and the replication proteins (RdRp).55
At this point, the global health community points to Gilead Sciences’ Remdesivir as the most effective existing drug that interferes with the virus’ replication process. Remdesivir was originally developed to treat Ebola and is a nucleoside analog, meaning that it is made up of synthetic RNA building blocks. When injected, the synthetic building blocks get in the way of viral RNA replication (interfering with RdRp) and make it difficult for the virus to make copies of itself. The drug currently has emergency FDA authorization and is in phase 3 clinical trials for treating COVID-19.56 Gilead recently announced that patients treated with the drug “were 65 percent more likely to have clinical improvement at Day 11” compared to those who were not.57
A number of new compounds are also under investigation. Emory University researchers’ EIDD-2801 antiviral compound works similarly to Remdesivir, can be ingested via pill, and is in phase 1 clinical trials.58 Other drugs could target the two viral protenases and nonstructural proteins like RNA helicase.59 In February, private biotech company Insilico used AI algorithms to find five new compounds that act as “protease inhibitors.”60 Alnylam, too, is working on a treatment that could inhibit viral SARS-CoV-2 replication through synthesized RNA strands called interference RNA (iRNA). This Nobel prize winning technique “silences” viral RdRp and RNA helicase.61
Limiting ability to harm: Finally, potential treatment options include drugs that limit the damage our own immune systems cause via inflammation and overreaction (cytokine storm) while fighting COVID-19. Many are investigating the efficacy of the existing drug, baricitinib, in targeting the AAK1 enzyme which can cause infection and inflammation in the lungs, one of COVID-19’s more lethal symptoms.62 Researchers are also investigating various arthritis drugs, which limit inflammatory responses by design, though results haven’t been positive so far.63
Vaccines
Vaccination efforts against SARS-CoV-2 seek to prime the immune system to fight off the virus before it can replicate and cause COVID-19. As of June 8, 2020, there were 161 vaccines in development around the world, with 10 in clinical trials.64,65
Vaccination-induced immunity is based on the fact that the virus neutralizing antibodies we produce when fighting pathogens stay in our systems for an extended period of time. This used to mainly entail injecting dead or weakened full pathogens (viruses) or fragments of them into our blood streams to induce an immune response. Or more technically, getting the B-cells in our immune systems to produce antibodies that bind to and block viral proteins called antigens.66 For context, SARS-CoV-2 antigens include the spike protein and other structural components. Several current efforts use this more traditional philosophy and include those that inject weakened/deactivated SARS-CoV-2 viruses, only antigens like the spike protein, and synthetic antigens.67
Others are looking to modern vaccine technology for a cure. Equipped with the virus’ genome, drug developers hope that they can take the genetic information that codes for SARS-CoV-2 antigens like the spike protein and make our bodies develop the antibodies that render it ineffective. Their thinking is that when injected, this genetic code can prompt our cells to produce safe, homegrown SARS-CoV-2 antigens which our immune systems would produce antibodies to “fight”– all without any exposure to the actual virus.68 There are multiple variations of this type of vaccine in development:
DNA-based vaccines consist of DNA sequences that code for antigens. The DNA enters our cells but requires a conversion (transcription) to RNA before its “instructions” can be read by cells’ ribosomes. Once transported into the nucleus, the foreign DNA utilizes the host cell’s transcription proteins for RNA conversion. The new RNA (from the injected DNA) then leaves the nucleus, and heads to ribosomes for antigen production.69
Compared to traditional vaccines, DNA vaccines present no risk of accidental viral infection, are more easily produced, and are safer for storage and shipping.70 These are key aspects given the immediate need to safely fight COVID-19 and as of June 8, 2020, there are 12 SARS-CoV-2 DNA vaccines in development. Of note, Takara Bio is working with a number of partners to produce a DNA vaccine for COVID-19. Phase 1 clinical trials should begin in July 2020.71
RNA-based vaccines inject RNA (rather than DNA) and share all of the same advantages of DNA vaccines, on top of some additional ones. Mainly, since our cells can immediately read RNA without conversion, RNA never has to enter our cells’ nuclei. This averts the risk of the antigen’s genetic material becoming part of our genome, and means smaller doses and traditional delivery mechanisms could be effective.72 DNA vaccines require high doses and unique delivery methods that can include electric shocks and the temporary insertion of metals under skin, while RNA vaccines are just like any shot you would get at the doctor’s office.73 As of June 8, 2020, there are 21 SARS-CoV-2 RNA-vaccines in development, with one candidate in phase 2.74
What’s Next for COVID-19 Treatments?
COVID-19 mobilized the global medical community to collaborate and pool resources to limit the devastating impacts and loss of life caused by the disease. Genomics enabled fast action in understanding the mechanisms that cause the disease, and while still in its early stages, could prove instrumental in treating symptoms and curing infections.
Genomics and a Healthier Future
COVID-19 is just one of many public health battles genomics seeks to remedy. While there is still much to learn about the human genome and how we can harness our knowledge of it, researchers and drug developers have made ground. Some genetic therapies already have approval to treat various chronic conditions. Sarepta Therapeutics’ FDA approved Vyondys 53, for example, treats Duchenne Muscular Dystrophy in patients with certain genetic traits.75
The gene therapy pipeline isn’t lagging. There are over 400 gene therapies in active clinical trials listed in the U.S. clinical trials database that span oncological uses, chronic conditions, infectious diseases, genetic diseases, and more.76 Editas Medicine and Allergan’s joint venture therapy to treat a certain kind of genetic blindness is currently in clinical trials.77 There are 34 clinical trials testing CRISPR gene-editing technology in treating a multitude of conditions: CRISPR therapeutics and Vertex Pharmaceuticals’ gene-editing therapy to treat sickle cell disease announced successful ½ phase clinical trials and the FDA recently gave it a Regenerative Medicine Advanced Therapy designation.78
Use cases and markets for genetic medicines could continue to expand and evolve as we learn more about the human genome, and ways to interact with and manipulate it.
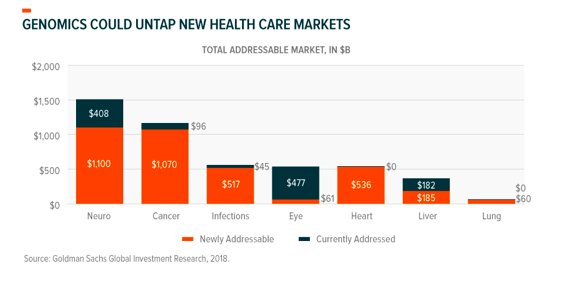
Beyond that, genomics is a totem for disruption in healthcare, ushering in an era of precision medicine that caters to individual needs and can optimize how doctors and organizations provide care. Genomics seeks to improve health now, could mean better health for future generations, and should help to prepare our world in facing the unknown. Investors looking for long-term, or even evergreen exposure to sector disruption should take notice.
Photo Credit: Yuri Samoilov via Flickr Creative Commons
The performance data quoted represents past performance and does not guarantee future results. Investment return and principal value of an investment will fluctuate so that an investor’s shares, when sold or redeemed, may be worth more or less than their original cost. Current performance may be higher or lower than the performance quoted. The fund recently experienced significant positive short-term performance due to market volatility associated with the COVID-19 pandemic. For performance data current to the most recent month end, please call 1-888-493-8631, or visit www.globalxetfs.com.
FOOTNOTES
1. Wu, Fan et al., Nature, “A new coronavirus associated with human respiratory disease in China,” 579, 265–269, February 3, 2020.
2. Zhang, Alison, Stanford University, “Sanger-vs-next-gen-sequencing,” July 2, 2019.
3. Chiu, Charles Y., Current Opinion in Microbiology, “Viral pathogen discovery,” 16(4), 468–478, May 29, 2013.
4. Ku, Chee-Seng et al., Expert Rev Med Devices, “From next-generation sequencing to nanopore sequencing technology: paving the way to personalized genomic medicine,”10(1), 1-6, January 9, 2014.
5. Chiu, Charles Y., Current Opinion in Microbiology, “Viral pathogen discovery,” 16(4), 468–478, May 29, 2013.
6. Zhu, Na et al., The New England Journal of Medicine, “A Novel Coronavirus from Patients with Pneumonia in China, 2019,” February 20, 2020.
7. BaseClear, “DNA/RNA sequencing Platforms: A short overview,” June 19, 2019.
8. Wu, Fan et al., NCBI,” Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome,” 579 (7798), 265-269, January 5, 2020.
9. Begley, Sharon, STAT, “DNA sleuths read the coronavirus genome, tracing its origins and looking for dangerous mutations,” January 24, 2020.
10. Colvin, Katherine, EMJ Microbiol Infect Dis. 2020, “Know Thine Enemy: Viral Genome Sequencing in Outbreaks,” 1(1), 34-37, June 1, 2020.
11. Oberholzer, Michael, Illumina, “Illumina Perspective on the Novel Coronavirus SARS-CoV-2 (2019-nCoV) Outbreak: What We Know Today and Where Do We Go from Here,” February 13, 2020.
12. IAEA, “How is the COVID-19 Virus Detected using Real Time RT-PCR?,” March 27, 2020.
13. S&P, “Hong Kong-listed Genscript Biotech ready to ramp up supply of coronavirus tests,” February 7, 2020.
14. Genscript, “What you need to know about SARS-CoV-2qPCR detection assay,” April 2, 2020.
15. Agilent, “Real Time PCR (qPCR) Instruments and software,” December 4, 2018.
16. Carter, Linda J. et al., ACS Central Science 2020, “Assay Techniques and Test Development for COVID-19 Diagnosis,” 6(5), 591–605, April 30, 2020.
17. Ibid.
18. Sethuraman, Nandini et al. JAMA Network, “Interpreting Diagnostic Tests for SARS-CoV-2,” 323(22), 2249-2251, May 6, 2020.
19. BioTek, “COVID-19: General Virology Applications,” 2020.
20. Yunes, Nadin et al., MDPI Viruses, “Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS-CoV-2,”
21. Krammer, Florian & Simon; Science, “Serology assays to manage COVID-19,” 368(6495), 1060-1061, June 5, 2020.
22. Ibid.
23. GenScript, “cPass™ SARS-CoV-2 Neutralization Antibody Detection Kit,” 2020.
24. Illumina, “An introduction to Next-Generation Sequencing Technology,”2020.
25. Gsaid.org
26. Zhou, Peng et al., Nature, “A pneumonia outbreak associated with a new coronavirus of probable bat origin,” 579, 270–273, February 3, 2020.
27. Ibid.
28. Kristian G. Anderson, et al. Nature Medicine, “The proximal origin of SARS-CoV-2,” 26, 450–452, March 17 2020
29. Nguyen, Thi et al., bioRxiv, “Origin of Novel Coronavirus (COVID-19): A Computational Biology Study using Artificial Intelligence,” Preprint, May 13, 2020.
30. Jarvis, Claire, Chemical & Engineering News, “How genomic epidemiology is tracking the spread of COVID-19 locally and globally,” 98(17), April 23, 2020.
31. Ibid.
32. Maurano, Matthew T. et al., Medrxiv, “Sequencing identifies multiple, early introductions of SARS-CoV2 to New York City Region,” Preprint, April 23, 2020.
33. Worobey, Michael et al.,Biorxiv, “The emergence of SARS-CoV-2 in Europe and the US,” Preprint, May 23, 2020.
34. Draghi, Jeremy & Ogbunu, Undark, “Opinion: The Coronavirus Is Mutating. That’s Not Necessarily Good or Bad,” May 14, 2020.
35. Our World in Data, 2020.
36. Milken Institute, “COVID-19 Treatment and Vaccine Tracker,” 2020.
37. FDA, “Emergency Use Authorization of Medical Products and Related Authorities,” October 17, 2018.
38. QIAGEN, “QIAGEN Ingenuity Pathway Analysis (QIAGEN IPA),” May 26, 2020.
39. Steve, Alexander et al., British Journal of Pharmacology, “A rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development,” Preprint, May 1, 2020.
40. IUPHAR/BPS Guide to Pharmacology, “SARS-CoV-2 Proteins,” 2020.
41. Hoffmann, Markus et al., Cell, “SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor,” 181(2), 731-733, April 16, 2020.
42. Gordon, David E. et al., Nature, “A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing,” Preprint, April 30, 2020.
43. Liu, Cynthia et al., ACS Central Science 2020, “Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases,” 6(3), 315-331, March 12, 2020.
44. Whitwam, Ryan, Extreme Tech, “IBM Supercomputer Identifies 77 Compounds That Could Fight Coronavirus,” March 20, 2020.
45. Sorrento Therapeutics, “COVID-19 Treatment: COVIDTRAP (ACE2 Receptor Decoy – STI 4398),” 2020.
46. SciNews, “New Antibody Completely Neutralizes SARS-CoV-2 Coronavirus,” May 18, 2020.
47. Salazar, Eric et al., The American Journal of Pathology, “Treatment of COVID-19 Patients with Convalescent Plasma,” Preprint, May 26, 2020.
48. Wrapp, Daniel et al., Cell, “Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies,” 181(5), 1004-1015, May 5, 2020.
49. Steve, Alexander et al., British Journal of Pharmacology, “A rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development,” Preprint, May 1, 2020.
50. Tomar, Sakshi et al., The Journal of Biological Chemistry, “Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro),” 290(32), 19403-19422, August 7, 2015.
51. Raj, V.S. et al., Current Opinion in Virology 2014, “MERS: emergence of a novel human coronavirus,” 5, 58-62, February 28, 2014.
52. Schmidtke, Peter, Discngine, “SARS-CoV-2 – part 2 – From the viral genome to protein structures,” March 27, 2020.
53. Chen, Yu et al., Journal of Medical Virology 2020, “Emerging coronaviruses: Genome structure, replication, and pathogenesis,” 92, 418-423, January 21, 2020.
54. Cascella, Marco et al., StatPearls Publishing, “Features, Evaluation and Treatment Coronavirus (COVID-19),” May 18, 2020.
55. Liu, Cynthia et al., ACS Central Science 2020, “Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases,” 6(3), 315-331, March 12, 2020.
56. Dolin, Raphael & Hirsch, The New England Journal of Medicine Editorials, “Remdesivir — An Important First Step,” May 27, 2020.
57. Gilead, “Gilead Announces Results From Phase 3 Trial of Remdesivir in Patients With Moderate COVID-19,” June 1, 2020.
58. Jarvis, Lisa M., Chemical & Engineering News, “Emory-discovered antiviral is poised for COVID-19 clinical trials,” 98(12), March 26, 2020.
59. Steve, Alexander et al., British Journal of Pharmacology, “A rational roadmap for SARS-CoV-2/COVID-19 pharmacotherapeutic research and development,” Preprint, May 1, 2020.
60. Lemonick, Sam, Chemical & Engineering News, “Two groups use artificial intelligence to find compounds that could fight the novel coronavirus” 98(12), February 4, 2020.
61. NBIC+, “RNA Interference Technology, Potential Treatment for Coronavirus Infection,” March 30, 2020.
62. Richardson, Peter et al., The Lancet Correspondence, “Baricitinib as potential treatment for 2019-nCoV acute respiratory disease,” 395(10223), 30-3, February 4, 2020.
63. Ibid.
64. Milken Institute, “COVID-19 Treatment and Vaccine Tracker,” 2020.
65. Mullard, Asher, The Lancet World Report, “COVID-19 vaccine development pipeline gears up,” 395(10239), 1751-1752, June 6, 2020.
66. Pacific Immunology, “How are antibodies produced,” 2020.
67. Liu, Cynthia et al., ACS Central Science 2020, “Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases,” 6(3), 315-331, March 12, 2020.
68. Liu, Margaret, Vaccines (Basel), “A Comparison of Plasmid DNA and mRNA as Vaccine Technologies,” 7(2), April 24, 2019.
69. Hobernik, Dominika & Bros, International Journal of Molecular Sciences, “DNA Vaccines—How Far From Clinical Use?,” 19(11), November 15, 2018.
70. Farris, Eriic et al., Experimental Biology & Medicine, “Micro- and nanoparticulates for DNA vaccine delivery,” 241(9), 919-929, April 4, 2016.
71. Akama, Kenya, Nikkei Asian Review, “Takara Bio drives Japan’s quest for a coronavirus vaccine,” May 27, 2020.
72. Verbeke, Rein et al., Nano Today 2019, “Three decades of messenger RNA vaccine development,” 28, October, 2019.
73. Moderna, mRNA, “Vaccines: Disruptive Innovation in Vaccination,” May, 2017.
74. Milken Institute, “COVID-19 Treatment and Vaccine Tracker,” 2020.
75. Sarepta, “Sarepta Therapeutics Announces FDA Approval of VYONDYS 53™ (golodirsen) Injection for the Treatment of Duchenne Muscular Dystrophy (DMD) in Patients Amenable to Skipping Exon 53,” December 12, 2019.
76. ClinicalTrials.Gov
77. The Motley Fool, “Why Editas Medicine Is Now the CRISPR Stock to Really Watch,” June 3, 2020.
78. CRISPR Therapeutics, “CRISPR Therapeutics and Vertex Pharmaceuticals Announce FDA Regenerative Medicine Advanced Therapy (RMAT) Designation Granted to CTX001™ for the Treatment of Severe Hemoglobinopathies,” May 11, 2019.



